Dagstuhl Seminar 24042
The Emerging Issues in Bioimaging AI Publications and Research
( Jan 21 – Jan 24, 2024 )
Permalink
Organizers
- Jianxu Chen (ISAS - Dortmund, DE)
- Florian Jug (Human Technopole - Milano, IT)
- Susanne Rafelski (Allen Insitute for Cell Science - Seattle, US)
- Shanghang Zhang (Peking University, CN)
Contact
- Marsha Kleinbauer (for scientific matters)
- Simone Schilke (for administrative matters)
Schedule
The seminar was divided into three specific directions: ethical considerations in bioimaging AI research and publications, performance reporting on bioimaging AI methods in publications and research, and future research directions of bioimaging AI focusing on validation and robustness. The seminar was structured into two parts: the first half focused on presentations and information sharing related to these three major directions to align experts from different fields, and the second half concentrated on in-depth discussions of these topics.
Given the highly interdisciplinary nature of the seminar, we took two specific steps to facilitate smooth communication and discussion among researchers with diverse backgrounds.
First, about six to eight weeks before the seminar, we sent out a survey to gather potential topics each participant could present within the seminar’s overarching theme. We collaborated with several participants to choose or adjust their presentation topics to ensure the effectiveness in this interdisciplinary setting. Based on the survey responses, the presentation and information-sharing portion (the first half) of our seminar began with two keynotes from editors who handle bioimaging AI papers, sharing their insights and the existing efforts by publishers. We then organized all presentations to progress from a focus on biology to bioimaging AI, and finally to AI, ensuring coverage of the full spectrum of necessary knowledge for our in-depth discussions in the second half.
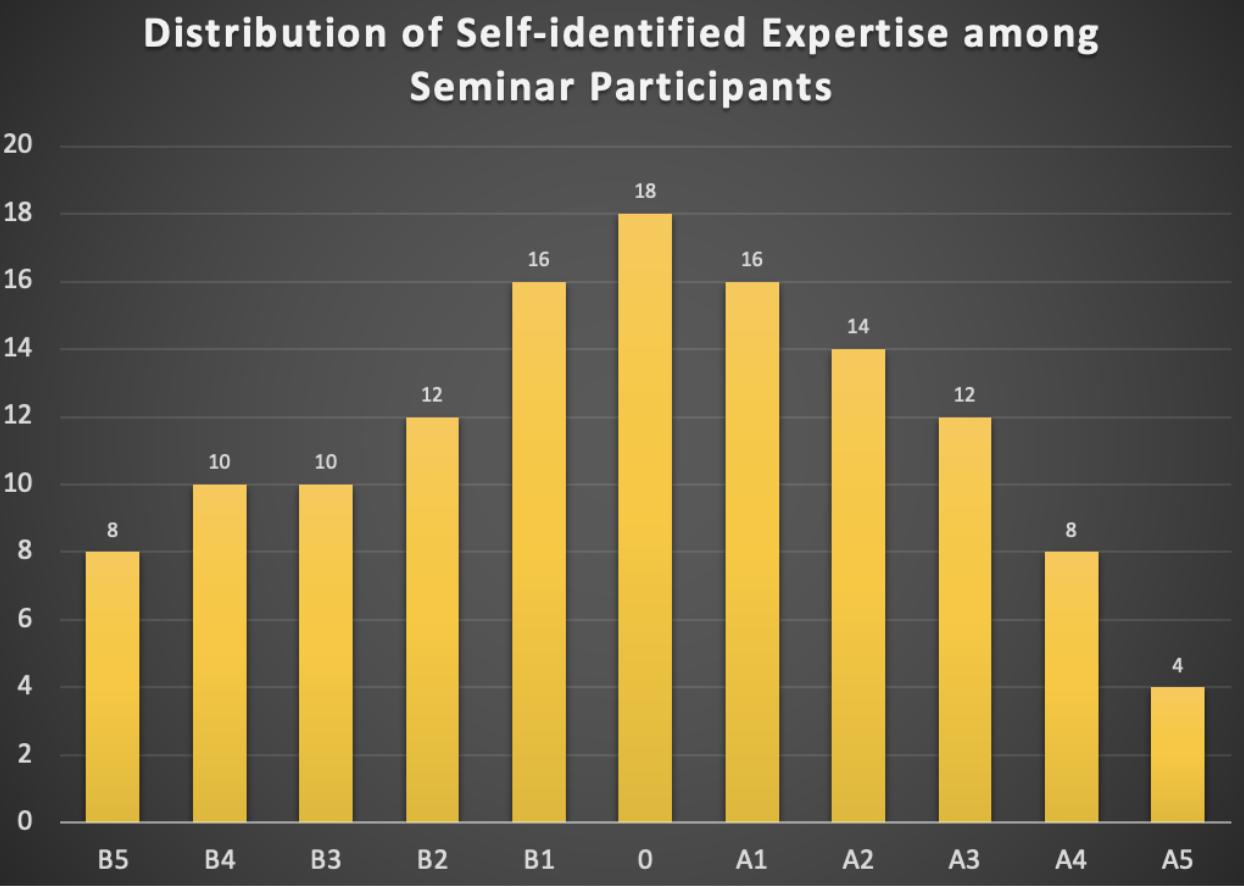
Second, at the beginning of the seminar, we allocated two minutes for each participant for a quick introduction and to briefly rate their experience and expertise on a scale in the range of [B5, B4, B3, B2, B1, 0, A1, A2, A3, A4, A5], with B5 representing pure biology and A5 representing pure AI. Participants could select a single value, multiple values, or a range of values. This was not intended to stereotype participants but to facilitate easier communication. For example, if a participant with experience in the range of B5 to B3 spoke with two others during a coffee break, one with experience from B3 to A1 and the other from A3 to A5, different communication strategies would be necessary for effective discussions. The distribution of self-identified experience is summarized in the histogram below (see Fig. 1).
Presentations, discussions and outcomes
Overview of the scientific talks
The seminar began with presentations by editors from Nature Methods and Cell Press, who shared their insights on existing and emerging issues in bioimaging AI publications. Following this, general bioimage analysis validation issues were discussed from both a biological application perspective and an algorithmic metric perspective. These presentations were succeeded by specific application talks demonstrating how AI-based bioimage analysis is utilized and validated in high-throughput biological applications [1]. The remainder of day one focused on bioimaging AI validation through explainable AI [2], [3], [4] and existing tools [5], as well as community efforts in deploying FAIR (Findable, Accessible, Interoperable, Reusable) AI tools for bioimage analysis [6].
The second day commenced with several theoretical AI talks introducing key concepts related to model robustness, fairness, and trustworthiness [7]. These were followed by two presentations showcasing state-of-the-art AI algorithms applied in bioimaging [8], [9], and an overview of the application of foundation models in bioimaging [10]. The scientific presentation portion of the seminar concluded with a talk about the pilot work initiated by the EMBO (European Molecular Biology Organization) Press on research integrity and AI integration in publishing and trust. This talk also served as a transition into the in-depth discussions that comprised the second part of the seminar.
Summary of discussions and key outcomes
After the scientific presentation part of the seminar, the participants naturally reach the agreement on doing the discussion in a four-quadrant manner, as illustrated below in Fig. 2.
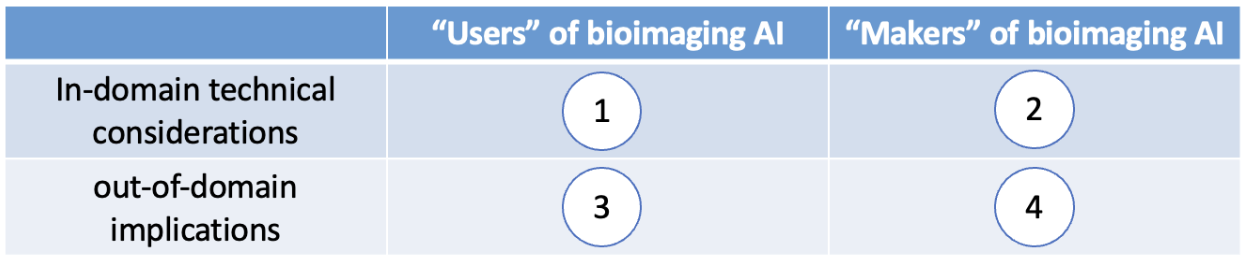
Here are some examples of what emerges from discussions in each quadrant.
I. What are some technical considerations that users of AI should pay attention to?
When using a specific bioimage analysis model, it is crucial for users to have clear biological questions that align with the technical limitations of the bioimaging AI models. This is known as application-appropriate validation [11]. For example, the trustworthiness or validity of an AI-based microscopy image denoising model may differ significantly between a study that requires merely counting the number of nuclei in an image and one that aims to quantify the morphological properties of the nuclei.
II. What are some technical considerations that makers of AI should pay attention
to?
When developing a bioimaging AI model, comprehensive evaluations and ablation studies are
essential to explicitly demonstrate the model’s limitations or potential failures. For instance,
evaluating a cell segmentation model under different conditions, such as various magnifications,
signal-to-noise ratios, cell densities, and possibly different microscope modalities, is highly beneficial. Providing a clear and detailed definition of the conditions under which the model
has been evaluated helps users determine whether the model can be directly applied to their
images or if it needs retraining or fine-tuning.
III. What are some important things the users of AI should make sure the makers
of AI are aware of or should make clear to the makers of AI?
One example is the inherent presence of batch effects in biology, such as variations in
fluorescence microscopy image quality due to different batches of dyes or slight morphological
differences in cells from different colony positions. For effective interdisciplinary collaboration,
it would be very helpful if biologists can clearly describe data acquisition processes and
potential batch effects. This enables AI developers to consider these factors in their training
sets, validation strategies, and model designs.
IV. What do the makers of AI need to make sure the users of AI know?
There is a lot of information that AI method developers need to help biologists think together.
For example, in some collaborative projects, AI researchers need to guide their biologist
collaborators how to best provide their data. For instance, the data to be analyzed to answer
biological questions can be different from special data acquired merely for training the AI
models, which could be referred to as a “training assay” [12], i.e., special experimental assays
only made for effective model training.
The discussions highlighted in the four quadrants are only examples from the seminar. A follow-up “white paper”-like manuscript based on the full discussions is being planned as a resource for the bioimaging AI community.
A specific topic that emerged from the seminar was the interpretability and explainability of bioimaging AI models. This is evident from the word cloud generated during the discussions, as shown in Fig. 3. A follow-up seminar specifically focusing on this topic is being planned for the bioimaging AI community in the coming years.

Besides the science program, the seminar provided valuable opportunities for social connections and networking. Due to the pandemic, many researchers had previously only met virtually, making the in-person interactions feel like a reunion. The diversity of research fields
Conclusions
This Dagstuhl Seminar on “The Emerging Issues in Bioimaging AI Publications and Research” successfully united a diverse group of experts from experimental biology, computational biology, bioimage analysis, computer vision, and AI research. The seminar facilitated indepth discussions on ethical considerations, performance reporting, and future research directions in bioimaging AI, highlighting the crucial need for interdisciplinary collaboration and communication.
Through structured presentations and interactive discussions, participants underscored the importance of clear communication between AI developers and users, comprehensive model validation, and awareness of biological batch effects. The seminar emphasized the necessity for application-appropriate validation and detailed reporting of AI model conditions to enhance the trustworthiness and applicability of bioimaging AI methods. Furthermore, the seminar provided a valuable platform for social interactions and networking, bridging gaps between researchers from different fields and fostering new collaborations.
In conclusion, the seminar not only advanced discussions on critical issues in bioimaging AI publications but also laid the foundation for ongoing collaboration and innovation in the field. Planned follow-up activities will further contribute to the development and ethical application of AI in bioimaging research. The success of this seminar underscores the importance of continuous communication and cooperation in addressing the emerging challenges in bioimaging AI publications and research.
Acknowledgement
We are grateful to all seminar participants for their insightful contributions and the engaging discussions they fostered, especially in the interdisciplinary setting with a wide spectrum of expertise. We also sincerely thank the Dagstuhl Scientific Directorate for the opportunity to organize this event. Finally, our deepest appreciation goes to the exceptional Dagstuhl staff whose support was instrumental in making the seminar a success.
References
- Z. Cibir et al., “ComplexEye: a multi-lens array microscope for high-throughput embedded immune cell migration analysis,” Nat. Commun., vol. 14, no. 1, p. 8103, Dec. 2023, doi: 10.1038/s41467-023-43765-3.
- Christopher J. Soelistyo and Alan R. Lowe, “Discovering Interpretable Models of Scientific Image Data with Deep Learning,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, 2024, pp. 6884–6893. [Online].
- Christopher J. Soelistyo, Guillaume Charras, and Alan R. Lowe, “Virtual Perturbations to Assess Explainability of Deep-Learning Based Cell Fate Predictors,” in Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) Workshops, 2023, pp. 3971–3980. [Online].
- D. Schuhmacher et al., “A framework for falsifiable explanations of machine learning models with an application in computational pathology,” Med. Image Anal., vol. 82, p. 102594, Nov. 2022, doi: 10.1016/j.media.2022.102594.
- L. M. Moser et al., “Piximi – An Images to Discovery web tool for bioimages and beyond.” Jun. 04, 2024. doi: 10.1101/2024.06.03.597232.
- W. Ouyang et al., “BioImage Model Zoo: A Community-Driven Resource for Accessible Deep Learning in BioImage Analysis,” Bioinformatics, preprint, Jun. 2022. doi: 10.1101/2022.06.07.495102.
- D. Guo, C. Wang, B. Wang, and H. Zha, “Learning Fair Representations via Distance Correlation Minimization,” IEEE Trans. Neural Netw. Learn. Syst., vol. 35, no. 2, pp. 2139–2152, Feb. 2024, doi: 10.1109/TNNLS.2022.3187165.
- Saumya Gupta, Yikai Zhang, Xiaoling Hu, and Prateek Prasanna, “Topology-aware uncertainty for image segmentation,” presented at the Advances in Neural Information Processing Systems, 2024.
- G. Dai et al., “Implicit Neural Image Field for Biological Microscopy Image Compression.” arXiv, 2024. doi: 10.48550/ARXIV.2405.19012.
- A. Archit et al., “Segment Anything for Microscopy,” Bioinformatics, preprint, Aug. 2023. doi: 10.1101/2023.08.21.554208.
- J. Chen, M. P. Viana, and S. M. Rafelski, “When seeing is not believing: applicationappropriate validation matters for quantitative bioimage analysis,” Nat. Methods, vol. 20, no. 7, pp. 968–970, Jul. 2023, doi: 10.1038/s41592-023-01881-4.
- J. Chen et al., “The Allen Cell and Structure Segmenter: a new open source toolkit for segmenting 3D intracellular structures in fluorescence microscopy images,” Cell Biology, preprint, Dec. 2018. doi: 10.1101/491035.
 Jianxu Chen, Florian Jug, Susanne Rafelski, and Shanghang Zhang
Jianxu Chen, Florian Jug, Susanne Rafelski, and Shanghang Zhang
The rapid development of artificial intelligence (AI) has revolutionized traditional microscopy imaging in many ways. Examples include ubiquitous tasks such as image denoising and restoration, image segmentation, object detection and classification, virtual staining, and more. While advances in all of these areas are being published in high-profile journals, this development has not yet been matched with standards and/or a community consensus on how to properly report results, conduct evaluations, and deal with ethical issues that come with technical AI advances. Hence, in this in-depth Dagstuhl Seminar, our discussions will be centered around two central topics, (i) the ethical use of AI in bioimaging and (ii) performance evaluation and reporting when using bioimage AI methods.
Who is invited to the seminar? We aim at bringing together people representing three types of expertise that all have an interest in finding solutions to the topics introduced above, namely, (i) AI researchers and bioimage AI method developers, (ii) users of AI facilitated methods and tools (life scientists, biologists, microscopists), and (iii) editors of journals handling bioimaging AI manuscripts.
What is the format of the seminar? Both main discussion topics will follow an “introduction and related works → brainstorming → outlook summary” format. We hope this seminar will serve as a kick-off meeting to initiate further community efforts on these and related topics.
For the topic of “ethical use of AI in bioimaging”, we first would like to raise the awareness that generative AI models have become so sophisticated that AI-generated bioimages can be indistinguishable from experimentally acquired images. Technically, we will discuss the state-of-the-art of image generation methods and how related AI fields deal with generated fake images (e.g., deepfake detection or image forgery detection). Then, the brainstorming session will aim to identify potential challenges in the bioimaging context and prospective solutions, followed, in the outlook session, by a discussion of potential research synergies and how community-driven guidelines on the ethical use of AI can be established.
For the topic of “AI performance evaluation and reporting”, we will first exchange insights about the status quo in bioimaging (e.g., common factors affecting microscopy image qualities, related biological applications, etc.) and then introduce the status quo on relevant topics in AI (e.g., domain shift, out-of-distribution detection, generalization gap, etc.). This will be followed up by introducing “application-appropriate validation” in bioimaging and possible evaluation strategies in related AI fields, such as medical imaging. The subsequent brainstorming session will begin with “case studies” from participants in order to understand and dissect the complexity and diversity of bioimaging evaluation problems, and then discuss potential solutions. Finally, the outlook session will focus on identifying future research synergies to address bottlenecks in bioimaging AI performance and the potential of establishing community standards for bioimaging AI performance evaluation and reporting.
What to expect at the end of the seminar? We expect all participants will gain a comprehensive overview of the demands, issues, potential solutions, and missing links to enable ethical, trustworthy, and publishable bioimaging AI methods, tools, and applications. We also aim to have this seminar kick off further meetings on these topics and create collaborations in the bioimaging AI community to follow up on ideas discussed during this event. Our hope is to follow up with participants online to transform the discussions in this seminar to an eventual joint community-driven perspective paper on these topics.
 Jianxu Chen, Florian Jug, and Susanne Rafelski
Jianxu Chen, Florian Jug, and Susanne Rafelski
- Chao Chen (Stony Brook University, US) [dblp]
- Jianxu Chen (ISAS - Dortmund, DE) [dblp]
- Evangelia Christodoulou (DKFZ - Heidelberg, DE)
- Beth Cimini (Broad Institute of MIT & Harvard - Cambridge, US) [dblp]
- Gaole Dai (Peking University, CN)
- Meghan Driscoll (University of Minnesota - Minneapolis, US) [dblp]
- Edward Evans III (University of Wisconsin - Madison, US)
- Matthias Gunzer (Universität Duisburg-Essen, DE & ISAS e.V. - Dortmund, DE)
- Andrew Hufton (Patterns, Cell Press - Würzburg, DE)
- Florian Jug (Human Technopole - Milano, IT) [dblp]
- Anna Kreshuk (EMBL - Heidelberg, DE) [dblp]
- Thomas Lemberger (EMBO - Heidelberg, DE)
- Alan Lowe (The Alan Turing Institute - London, GB)
- Shalin Mehta (Chan Zuckerberg Biohub - Stanford, US)
- Axel Mosig (Ruhr-Universität Bochum, DE) [dblp]
- Matheus Palhares Viana (Allen Insitute for Cell Science - Seattle, US)
- Constantin Pape (Universität Göttingen, DE) [dblp]
- Anne Plant (NIST - Gaithersburg, US)
- Susanne Rafelski (Allen Insitute for Cell Science - Seattle, US) [dblp]
- Ananya Rastogi (Springer Nature - New York, US)
- Albert Sickmann (ISAS - Dortmund, DE) [dblp]
- Rita Strack (Nature Publishing Group, US)
- Nicola Strisciuglio (University of Twente - Enschede, NL)
- Aubrey Weigel (Howard Hughes Medical Institute - Ashburn, US)
- Assaf Zaritsky (Ben Gurion University - Beer Sheva, IL) [dblp]
- Shanghang Zhang (Peking University, CN) [dblp]
- Han Zhao (University of Illinois - Urbana-Champaign, US) [dblp]
Classification
- Artificial Intelligence
- Computer Vision and Pattern Recognition
- Machine Learning
Keywords
- artificial intelligence
- publication ethics
- bioimaging
- trustworthy AI
- open source




 Creative Commons BY 4.0
Creative Commons BY 4.0
